Product Cat. No.: GBS-010-1
For Research Use Only.
ERG, TMPRSS, ETV1, ETV4 gene anomaly probe detection kit
10 Tests/box
This kit uses fluorescence in situ hybridization to detect the gene status of ERG, TMPRSS2, ETV1, and ETV4 in vitro. The test samples are paraffin embedded tissue samples from suspected or diagnosed prostate cancer patients.
Prostate cancer is a common malignant tumor in men. In recent years, the incidence of prostate cancer in China has shown an obvious upward trend. About 80% of prostate cancer patients are accompanied by genetic changes of the fusion of TMPRSS2 gene and ETS gene family (such as ERG, ETV1, ETV4), but these changes do not occur in benign prostate diseases. FISH testing can provide more accurate and objective diagnostic indicators for clinical use compared to PSA screening and biopsy.
This kit has only been validated for the detection performance of ERG, TMPRSS2, ETV1, and ETV4 genes. This kit is only suitable for detecting the gene status of ERG, TMPRSS2, ETV1, and ETV4, providing diagnostic assistance for physicians.
Fluorescence in situ hybridization is a technique that directly observes = specific nucleic acids in cells in vitro. According to the principle of complementary base pairing, a specific DNA sequence complements and binds to the target sequence within the cell. Due to the fluorescence of the probe, under appropriate excitation light irradiation, the hybrid probe and target DNA can be clearly observed under a fluorescence microscope.
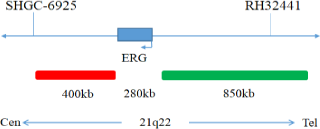
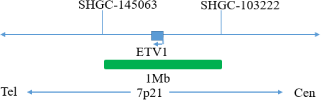
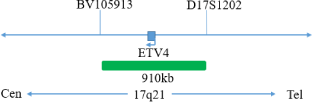
This kit uses orange fluorescent labeled orange red probes(ERG gene 5 ‘end), TMPRSS2 probes, and green fluorescent labeled green probes (ERG gene 3’ end), ETV1, and ETV4 probesto bind the two probes to the target detection site through in situ hybridization technology. When ERG gene recombination occurs, the ERG dual color probe signal separation appears as a distant monochromatic signal. When TMPRSS2 gene and ETV1 gene fuse, the TMPRSS2/ETV1 dual color probe signal fuses to form a yellow fusion signal. When TMPRSS2 gene and ETV4 gene fuse, the TMPRSS2/ETV4 dual color probe signal fuses to form a yellow fusion signal.
The kit consists of one of TMPRSS2/ETV1, TMPRSS2/ETV4, ERG dual color probes, as shown in Table 1.
| Cat# | Component name | Specifications | Quantity | Main components |
|---|---|---|---|---|
| GBS-010-1 | ERG dual color probe | 100μL/Tube | 1 | ERG Orange probe; ERG Green probe |
| GBS-010-2 | TMPRSS2/ETV1 dual color probe | 100μL/Tube | 1 | TMPRSS2 Orange probe; ETV1 Green probe |
| GBS-010-3 | TMPRSS2/ETV4 dual color probe | 100μL/Tube | 1 | TMPRSS2 Orange probe; ETV4 Green probe |

Keep sealed away from light at -20°C±5°C. The product is valid for 20 months. Avoid unnecessary repeated freezing and thawing that should not exceed 10 times. After opening, within 24 hours for short-term preservation, keep sealed at 2~8°C in dark. For long-term preservation after opening, keep the lid sealed at -20°C±5°C away from light. The kit is transported below 0°C.
Fluorescence microscopy imaging systems including fluorescence microscopy and filter sets suitable for DAPI, Green, and Orange.
1. Applicable specimen type: Paraffin embedded tissue samples from suspected or diagnosed prostate cancer patients.
2. The specimen should be fixed with 4% neutral formalin fixation solution within 1 hour after being detached, and the specimen should be regularly dehydrated and paraffin embedded after fixation.
3. The thickness of paraffin slices will affect the experimental results, with a slice thickness of 4-5μm is appropriate.
4. Representative tumor tissue wax blocks should be selected from paraffin embedded tissue specimens and confirmed by HE staining.
5. It is recommended to choose paraffin embedded tissue specimens with a shorter storage time (within 5 years).
Sodium chloride
176g
Sodium citrate
88g
Weigh 176g of sodium chloride and 88g of sodium citrate, dissolve in 800mL of deionized water, adjust the pH to 5.3±0.2 at room temperature, and complete to 1 L with deionized water. High-pressure steam sterilization, stored at 2~8°C, the solution shelf life is of 6 months. Discard if the reagent appears cloudy (turbid) or contaminated.
NP-40
0.6mL
20xSSC
4mL
Take 0.6mL NP-40 and 4mL 20×SSC, add 150mL deionized water, mix, adjust the pH to 7.0 ~ 7.5 at room temperature, with deionized water complete to a volume of 200mL. Stored at 2~8°C, the shelf life is of 6 months. Discard if the reagent appears cloudy (turbid) or contaminated.
1. Signal classification and counting
2. FISH result judgment
To determine the anomalies in the detection results, it is necessary to establish an anomaly threshold
This kit is used for paraffin embedded specimens of prostate cancer tissue and is not recommended for use in other tissues. The detection ability of paraffin tissue samples that have been stored for too long cannot be evaluated according to this instruction; Follow the procedures provided in this manual, as changing the procedures may alter the results of the inspection; This kit only detects the gene status of ERG,TMPRSS2, ETV1, and ETV4, and cannot be used as the sole basis for diagnosis, prognosis judgment, or other clinical management of prostate cancer patients. A comprehensive evaluation should be conducted based on the patient’s medical history and other diagnostic results.
1. Fluorescence signal intensity: After effective hybridization with the karyotype reference material, the probe should emit a fluorescence signal that can be recognized by the naked eye under a fluorescence microscope.
2. Effective rate: Three negative reference samples and three positive reference samples were tested, and the accuracy rate of the results was 100%.
3. Sensitivity:
3.1 Sensitivity of TMPRSS2 gene probe
After effective hybridization with karyotype reference materials, the probe analyzed 100 chromosomes 21 from 50 cells in the metaphase division phase, and at least 98 chromosomes 21 showed an orange red fluorescence signal.
3.2 ETV1 gene probe sensitivity
After effective hybridization with karyotype reference materials, the probe analyzed 100 chromosomes 7 from 50 cells in the metaphase division phase, and at least 98 chromosomes 7 showed 1 green fluorescence signal.
3.3 ETV4 gene probe sensitivity
After effective hybridization with karyotype reference materials, the probe analyzed 100 chromosomes 17 from 50 cells in the metaphase division phase, and at least 98 chromosomes 17 showed 1 green fluorescence signal.
3.4 ERG Orange Red Probe Sensitivity
After effective hybridization with karyotype reference materials, the probe analyzed 100 chromosomes 21 from 50 cells in the metaphase division phase, and at least 98 chromosomes 21 showed an orange red fluorescence signal.
3.5 ERG Green Probe Sensitivity
After effective hybridization with karyotype reference materials, the probe analyzed 100 chromosomes 21 from 50 cells in the metaphase division phase, and at least 98 chromosomes 21 showed 1 green fluorescence signal.
4. Specificity:
4.1 TMPRSS2 gene probe specificity
After effective hybridization with karyotype reference materials, the probe analyzed 100 chromosomes 21 from 50 cells in the metaphase division phase, and at least 98 chromosomes 21 displayed a specific orange red fluorescence signal in the long arm target area.
4.2 ETV1 gene probe specificity
After effective hybridization with karyotype reference materials, the probe analyzed 100 chromosomes 7 from 50 cells in the metaphase division phase, and at least 98 chromosomes 7 displayed a specific green fluorescence signal in the short arm target region.
4.3 ETV4 gene probe specificity
After effective hybridization with karyotype reference materials, the probe analyzed 100 chromosomes 17 from 50 cells in the metaphase division phase, and at least 98 chromosomes 17 displayed a specific green fluorescence signal in the long arm target area.
4.4 ERG orange probe specificity
After effective hybridization with karyotype reference materials, the probe analyzed 100 chromosomes 21 from 50 cells in the metaphase division phase, and at least 98 chromosomes 21 displayed a specific orange red fluorescence signal in the long arm target area.
4.5 ERG green probe specificity
After effective hybridization with karyotype reference materials, the probe analyzed 100 chromosomes 21 from 50 cells in the metaphase division phase, and at least 98 chromosomes 21 displayed a specific green fluorescence signal in the target region.
1. Please read this manual carefully before testing. Testing personnel should receive professional technical training, and signal counting personnel must be able to observe and distinguish orange red and green signals.
2.When testing clinical samples, when the hybridization signal count is difficult and the sample is not sufficient to repeat the retest, the test will not provide any test results. If the cell count is insufficient for analysis, the test will also not provide test results.
3. The DAPI dye used in this experiment has potential toxicity or carcinogenicity, and should be operated in a fume hood, wearing masks and gloves to avoid direct contact.
4. The results of this reagent kit may be affected by various factors within the sample itself, as well as limitations such as enzyme digestion time, hybridization temperature and time, operating environment, and limitations of current molecular biology technology, which may lead to incorrect interpretation results. Users must understand the potential errors and limitations of accuracy that may exist during the testing process.
5. All chemicals have potential hazards and should be avoided from direct contact. Used reagent kits are clinical waste and should be properly disposed of.
V1. 0: Approval date: May 08, 2019.
V1. 4: Revision date: December 07, 2021.
